
Illuminating The Druggable Genome Resource Dissemination And Outreach Center

Stephan C. Schürer, Ph.D.

Professor, Department of Pharmacology, Miller School of Medicine, University of Miami
Associate Director of Data Science, Sylvester Comprehensive Cancer Center, Miller School of Medicine, University of Miami
Director of Digital Drug Discovery, Institute for Data Science & Computing, University of Miami
University of Miami
Associate Director of Data Science, Sylvester Comprehensive Cancer Center, Miller School of Medicine, University of Miami
Director of Digital Drug Discovery, Institute for Data Science & Computing, University of Miami

Overview
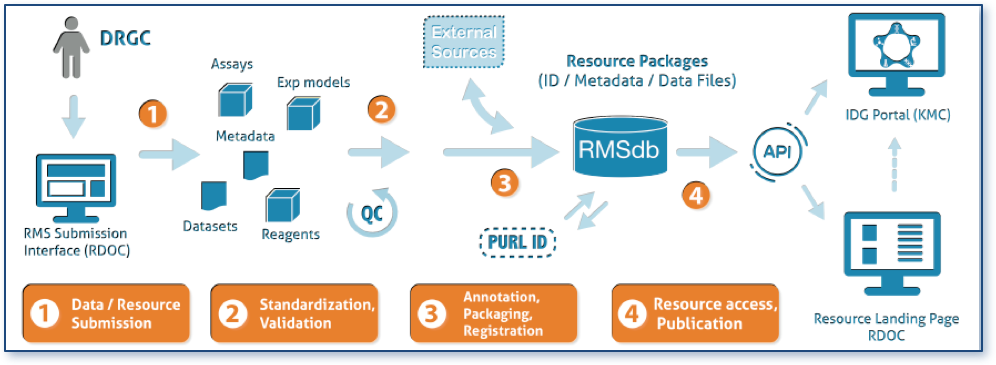
The RDOC grant at U of Miami is led by Dr. Stephan Schürer. Work from his group will focus on dissemination of the resources generated from the DRGC by stewarding implementation of metadata standards for IDG and developing a Resource Management System (RMS) capable of a variety of data and resources.
NIH grant number: 1U24TR002278-01
RDOC at University of Miami publications:
- Vidović D, Waller A, Holmes J, Sklar LA, Schürer SC. Best practices for managing and disseminating resources and outreach and evaluating the impact of the IDG Consortium. Drug Discov. Today 2024 Mar 18. doi: 10.1016/j.drudis.2024.103953. PMID: 38508231
- Issa NT, Stathias V, Schürer S, Dakshanamurthy S. Machine and deep learning approaches for cancer drug repurposing. Semin Cancer Biol. 2021 Jan;68:132-142. doi: 10.1016/j.semcancer.2019.12.011. Epub 2020 Jan 3. PMID: 31904426; PMCID: PMC7723306.
- Chaudagar K, Hieromnimon HM, Khurana R, Labadie B, Hirz T, Mei S, Hasan R, Shafran J, Kelley A, Apostolov E, Al-Eryani G, Harvey K, Rameshbabu S, Loyd M, Bynoe K, Drovetsky C, Solanki A, Markiewicz E, Zamora M, Fan X, Schürer S, Swarbrick A, Sykes DB, Patnaik A. Reversal of Lactate and PD-1-mediated Macrophage Immunosuppression Controls Growth of PTEN/p53-deficient Prostate Cancer. Clin Cancer Res. 2023 May 15;29(10):1952-1968. doi: 10.1158/1078-0432.CCR-22-3350. PMID: 36862086; PMCID: PMC10192075.
- Ursu O, Holmes J, Bologa CG, Yang JJ, Mathias SL, Stathias V, Nguyen DT, Schürer S, Oprea T. DrugCentral 2018: an update. Nucleic Acids Res. 2019 Jan 8;47(D1):D963-D970. doi: 10.1093/nar/gky963. PMID: 30371892; PMCID: PMC6323925.
- Oprea TI, Bologa CG, Brunak S, Campbell A, Gan GN, Gaulton A, Gomez SM, Guha R, Hersey A, Holmes J, Jadhav A, Jensen LJ, Johnson GL, Karlson A, Leach AR, Ma'ayan A, Malovannaya A, Mani S, Mathias SL, McManus MT, Meehan TF, von Mering C, Muthas D, Nguyen DT, Overington JP, Papadatos G, Qin J, Reich C, Roth BL, Schürer SC, Simeonov A, Sklar LA, Southall N, Tomita S, Tudose I, Ursu O, Vidovic D, Waller A, Westergaard D, Yang JJ, Zahoránszky-Köhalmi G. Unexplored therapeutic opportunities in the human genome. Nat Rev Drug Discov. 2018 May;17(5):317-332. doi: 10.1038/nrd.2018.14. Epub 2018 Mar 23. Erratum in: Nat Rev Drug Discov. 2018 Mar 23;: PMID: 29472638; PMCID: PMC6339563.
- Kelleher KJ, Sheils TK, Mathias SL, Yang JJ, Metzger VT, Siramshetty VB, Nguyen DT, Jensen LJ, Vidović D, Schürer SC, Holmes J, Sharma KR, Pillai A, Bologa CG, Edwards JS, Mathé EA, Oprea TI. Pharos 2023: an integrated resource for the understudied human proteome. Nucleic Acids Res. 2023 Jan 6;51(D1):D1405-D1416. doi: 10.1093/nar/gkac1033. Erratum in: Nucleic Acids Res. 2023 Feb 28;51(4):1999. PMID: 36624666; PMCID: PMC9825581.
- Muratov EN , Amaro R , Andrade CH , Brown N , Ekins S , Fourches D , Isayev O , Kozakov D , Medina-Franco JL , Merz KM , Oprea TI , Poroikov V , Schneider G , Todd MH , Varnek A , Winkler DA , Zakharov AV , Cherkasov A , Tropsha A . A critical overview of computational approaches employed for COVID-19 drug discovery. Chem Soc Rev. 2021 Aug 21;50(16):9121-9151. doi: 10.1039/d0cs01065k. Epub 2021 Jul 2. PMID: 34212944; PMCID: PMC8371861.
- Issa NT, Badiavas EV, Schürer S. Research Techniques Made Simple: Molecular Docking in Dermatology - A Foray into In Silico Drug Discovery. J Invest Dermatol. 2019 Dec;139(12):2400-2408.e1. doi: 10.1016/j.jid.2019.06.129. PMID: 31753122.
- Sheils TK, Mathias SL, Kelleher KJ, Siramshetty VB, Nguyen DT, Bologa CG, Jensen LJ, Vidović D, Koleti A, Schürer SC, Waller A, Yang JJ, Holmes J, Bocci G, Southall N, Dharkar P, Mathé E, Simeonov A, Oprea TI. TCRD and Pharos 2021: mining the human proteome for disease biology. Nucleic Acids Res. 2021 Jan 8;49(D1):D1334-D1346. doi: 10.1093/nar/gkaa993. PMID: 33156327; PMCID: PMC7778974.
- Stathias V, Turner J, Koleti A, Vidovic D, Cooper D, Fazel-Najafabadi M, Pilarczyk M, Terryn R, Chung C, Umeano A, Clarke DJB, Lachmann A, Evangelista JE, Ma'ayan A, Medvedovic M, Schürer SC. LINCS Data Portal 2.0: next generation access point for perturbation-response signatures. Nucleic Acids Res. 2020 Jan 8;48(D1):D431-D439. doi: 10.1093/nar/gkz1023. PMID: 31701147; PMCID: PMC7145650.
- Cichońska A, Ravikumar B, Allaway RJ, Wan F, Park S, Isayev O, Li S, Mason M, Lamb A, Tanoli Z, Jeon M, Kim S, Popova M, Capuzzi S, Zeng J, Dang K, Koytiger G, Kang J, Wells CI, Willson TM; IDG-DREAM Drug-Kinase Binding Prediction Challenge Consortium; Oprea TI, Schlessinger A, Drewry DH, Stolovitzky G, Wennerberg K, Guinney J, Aittokallio T. Crowdsourced mapping of unexplored target space of kinase inhibitors. Nat Commun. 2021 Jun 3;12(1):3307. doi: 10.1038/s41467-021-23165-1. PMID: 34083538; PMCID: PMC8175708.
- Essegian DJ, Chavez V, Bustamante F, Schürer SC, Merchan JR. Cellular and molecular effects of PNCK, a non-canonical kinase target in renal cell carcinoma. iScience. 2022 Nov 17;25(12):105621. doi: 10.1016/j.isci.2022.105621. PMID: 36465101; PMCID: PMC9713373.
- Kropiwnicki E, Binder JL, Yang JJ, Holmes J, Lachmann A, Clarke DJB, Sheils T, Kelleher KJ, Metzger VT, Bologa CG, Oprea TI, Ma'ayan A. Getting Started with the IDG KMC Datasets and Tools. Curr Protoc. 2022 Jan;2(1):e355. doi: 10.1002/cpz1.355. PMID: 35085427; PMCID: PMC10789444.
- Ackloo S, Al-Awar R, Amaro RE, Arrowsmith CH, Azevedo H, Batey RA, Bengio Y, Betz UAK, Bologa CG, Chodera JD, Cornell WD, Dunham I, Ecker GF, Edfeldt K, Edwards AM, Gilson MK, Gordijo CR, Hessler G, Hillisch A, Hogner A, Irwin JJ, Jansen JM, Kuhn D, Leach AR, Lee AA, Lessel U, Morgan MR, Moult J, Muegge I, Oprea TI, Perry BG, Riley P, Rousseaux SAL, Saikatendu KS, Santhakumar V, Schapira M, Scholten C, Todd MH, Vedadi M, Volkamer A, Willson TM. CACHE (Critical Assessment of Computational Hit-finding Experiments): A public-private partnership benchmarking initiative to enable the development of computational methods for hit-finding. Nat Rev Chem. 2022 Apr;6(4):287-295. doi: 10.1038/s41570-022-00363-z. Epub 2022 Feb 15. PMID: 35783295; PMCID: PMC9246350.
- Cooper DJ, Schürer S. Improving the Utility of the Tox21 Dataset by Deep Metadata Annotations and Constructing Reusable Benchmarked Chemical Reference Signatures. Molecules. 2019 Apr 23;24(8):1604. doi: 10.3390/molecules24081604. PMID: 31018579; PMCID: PMC6515292.
- Küçük McGinty H, Visser U, Schürer S. How to Develop a Drug Target Ontology: KNowledge Acquisition and Representation Methodology (KNARM). Methods Mol Biol. 2019;1939:49-69. doi: 10.1007/978-1-4939-9089-4_4. PMID: 30848456; PMCID: PMC7257161.
- He Y, Duncan WD, Cooper DJ, Hansen J, Iyengar R, Ong E, Walker K, Tibi O, Smith S, Serra LM, Zheng J, Sarntivijai S, Schürer S, O'Shea KS, Diehl AD. OSCI: standardized stem cell ontology representation and use cases for stem cell investigation. BMC Bioinformatics. 2019 Apr 25;20(Suppl 5):180. doi: 10.1186/s12859-019-2723-7. PMID: 31272389; PMCID: PMC6509805.
- Stathias V, Jermakowicz AM, Maloof ME, Forlin M, Walters W, Suter RK, Durante MA, Williams SL, Harbour JW, Volmar CH, Lyons NJ, Wahlestedt C, Graham RM, Ivan ME, Komotar RJ, Sarkaria JN, Subramanian A, Golub TR, Schürer SC, Ayad NG. Drug and disease signature integration identifies synergistic combinations in glioblastoma. Nat Commun. 2018 Dec 14;9(1):5315. doi: 10.1038/s41467-018-07659-z. PMID: 30552330; PMCID: PMC6294341.
- Oprea TI, Bologa CG, Brunak S, Campbell A, Gan GN, Gaulton A, Gomez SM, Guha R, Hersey A, Holmes J, Jadhav A, Jensen LJ, Johnson GL, Karlson A, Leach AR, Ma'ayan A, Malovannaya A, Mani S, Mathias SL, McManus MT, Meehan TF, von Mering C, Muthas D, Nguyen DT, Overington JP, Papadatos G, Qin J, Reich C, Roth BL, Schürer SC, Simeonov A, Sklar LA, Southall N, Tomita S, Tudose I, Ursu O, Vidovic D, Waller A, Westergaard D, Yang JJ, Zahoránszky-Köhalmi G. Unexplored therapeutic opportunities in the human genome. Nat Rev Drug Discov. 2018 May;17(5):377. doi: 10.1038/nrd.2018.52. Epub 2018 Mar 23. Erratum for: Nat Rev Drug Discov. 2018 May;17 (5):317-332. PMID: 29567993.
- Essegian DJ, Chavez V, Khurshid R, Merchan JR, Schürer SC. AI-Assisted chemical probe discovery for the understudied Calcium-Calmodulin Dependent Kinase, PNCK. PLoS Comput Biol. 2023 May 26;19(5):e1010263. doi: 10.1371/journal.pcbi.1010263. Erratum in: PLoS Comput Biol. 2023 Nov 22;19(11):e1011672. PMID: 37235579; PMCID: PMC10249896.
- Gibbons GS, Chakraborty A, Grigsby SM, Umeano AC, Liao C, Moukha-Chafiq O, Pathak V, Mathew B, Lee YT, Dou Y, Schürer SC, Reynolds RC, Snowden TS, Nikolovska-Coleska Z. Identification of DOT1L inhibitors by structure-based virtual screening adapted from a nucleoside-focused library. Eur J Med Chem. 2020 Mar 1;189:112023. doi: 10.1016/j.ejmech.2019.112023. Epub 2020 Jan 2. PMID: 31978781; PMCID: PMC7646624.
- Essegian D, Khurana R, Stathias V, Schürer SC. The Clinical Kinase Index: A Method to Prioritize Understudied Kinases as Drug Targets for the Treatment of Cancer. Cell Rep Med. 2020 Oct 20;1(7):100128. doi: 10.1016/j.xcrm.2020.100128. PMID: 33205077; PMCID: PMC7659504.
Contacts
- Stephan Schurer’s lab: ccs.miami.edu/
- Drug Target Ontology: http://drugtargetontology.org
- Drug Discovery lab: https://idsc.miami.edu/research/digital-health/drug-discovery/
