
Knowledge Management Center for Illuminating the Druggable Genome

Avi Ma’ayan, PhD
Principal Investigator
Professor, Department of Pharmacological Sciences
Director, Mount Sinai Center for Bioinformatics
Icahn School of Medicine at Mount Sinai
Overview
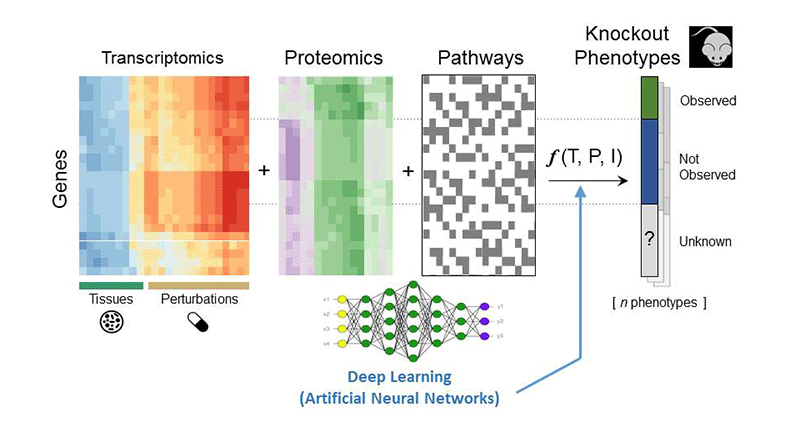
To better understand the function of the understudied protein targets, which are the focus of the implementation phase of the Illuminating the Druggable Genome (IDG) project, we impute knowledge using machine learning strategies. To establish this classification system, we organize data from many omics- and literature-based resources into attribute tables where genes are the rows and their attributes are the columns. Examples of such attribute tables include gene or protein expression in cancer cell lines (CCLE) or human tissues (GTEx), changes in expression in response to drug perturbations or single-gene knockdowns (LINCS), regulation by transcription factors based on ChIP-seq data (ENCODE), and phenotypes in mice observed when single genes are knocked out (KOMP). In total, we process and abstract data from over 100 resources. We then predict target function, target association with pathways, small-molecules/drugs that modulate the activity and expression of the target, and target relevance to human disease. Overall, the KMC-ISMMS develops a useful resource that will accelerate target and drug discovery.
NIH grant number: U24CA224260-01
Web
- Ma’ayan Laboratory: https://labs.icahn.mssm.edu/maayanlab/
- Mount Sinai Center for Bioinformatics: https://icahn.mssm.edu/research/bioinformatics
- Harmonizome https://maayanlab.cloud/Harmonizome/
- Geneshot https://amp.pharm.mssm.edu/geneshot/
- CREEDS https://amp.pharm.mssm.edu/CREEDS/
- Enrichr https://maayanlab.cloud/Enrichr/
- Clustergrammer https://maayanlab.cloud/clustergrammer/
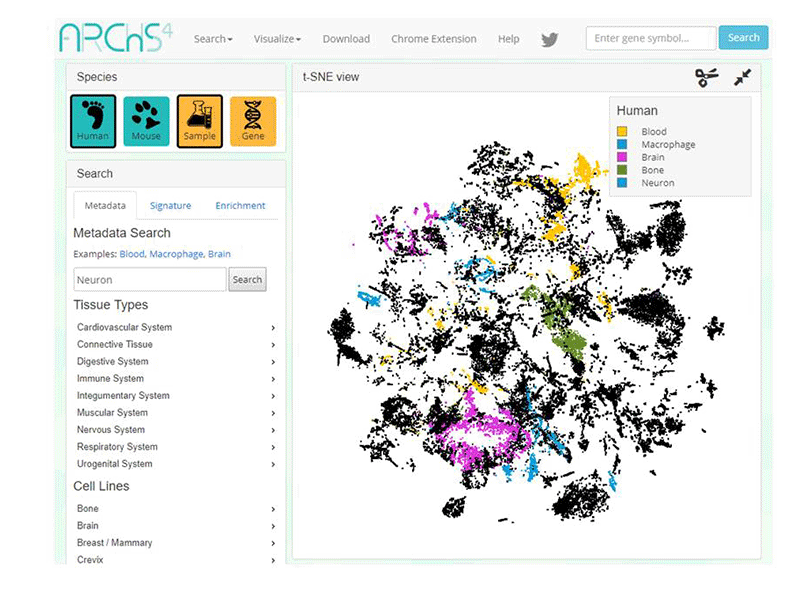
- ARCHS4 https://amp.pharm.mssm.edu/archs4
You Tube
https://www.youtube.com/user/avimaayan
https://www.linkedin.com/in/avi-ma-ayan-6a61271a/
KMC-ISMMS publications:
- Deng EZ, Marino GB, Clarke DJB, Diamant I, Resnick AC, Ma W, Wang P, Ma'ayan A. Multiomics2Targets identifies targets from cancer cohorts profiled with transcriptomics, proteomics, and phosphoproteomics. Cell Rep Methods. 2024 Aug 4:100839. doi: 10.1016/j.crmeth.2024.100839. Online ahead of print. PMID: 39127042
- Clarke DJB, Marino GB, Deng EZ, Xie Z, Evangelista JE, Ma'ayan A. Rummagene: massive mining of gene sets from supporting materials of biomedical research publications. Commun Biol. 2024 Apr 20;7(1):482. doi: 10.1038/s42003-024-06177-7. PMID: 38643247
- Xie Z, Chen C, Ma'ayan A. Dex-Benchmark: datasets and code to evaluate algorithms for transcriptomics data analysis. PeerJ. 2023 Nov 8;11:e16351. doi: 10.7717/peerj.16351. PMID: 37953774; PMCID: PMC10638921.
- Evangelista JE, Xie Z, Marino GB, Nguyen N, Clarke DJB, Ma'ayan A. Enrichr- KG: bridging enrichment analysis across multiple libraries. Nucleic Acids Res. 2023 Jul 5;51(W1):W168-W179. doi: 10.1093/nar/gkad393. PMID: 37166973; PMCID: PMC10320098.
- Marino GB, Ngai M, Clarke DJB, Fleishman RH, Deng EZ, Xie Z, Ahmed N, Ma'ayan A. GeneRanger and TargetRanger: processed gene and protein expression levels across cells and tissues for target discovery. Nucleic Acids Res. 2023 Jul 5;51(W1):W213-W224. doi: 10.1093/nar/gkad399. PMID: 37166966; PMCID: PMC10320068.
- Deng EZ, Fleishman RH, Xie Z, Marino GB, Clarke DJB, Ma'ayan A. Computational screen to identify potential targets for immunotherapeutic identification and removal of senescence cells. Aging Cell. 2023 Jun;22(6):e13809. doi: 10.1111/acel.13809. Epub 2023 Apr 20. PMID: 37082798; PMCID: PMC10265163.
- Lachmann A, Rizzo KA, Bartal A, Jeon M, Clarke DJB, Ma'ayan A. PrismEXP: gene annotation prediction from stratified gene-gene co-expression matrices. PeerJ. 2023 Feb 27;11:e14927. doi: 10.7717/peerj.14927. PMID: 36874981; PMCID: PMC9979837.
- Marino GB, Wojciechowicz ML, Clarke DJB, Kuleshov MV, Xie Z, Jeon M, Lachmann A, Ma'ayan A. lncHUB2: aggregated and inferred knowledge about human and mouse lncRNAs. Database (Oxford). 2023 Mar 4;2023:baad009. doi: 10.1093/database/baad009. PMID: 36869839; PMCID: PMC9985331.
- Jeon M, Xie Z, Evangelista JE, Wojciechowicz ML, Clarke DJB, Ma'ayan A. Transforming L1000 profiles to RNA-seq-like profiles with deep learning. BMC Bioinformatics. 2022 Sep 13;23(1):374. doi: 10.1186/s12859-022-04895-5. PMID: 36100892; PMCID: PMC9472394.
- Xie Z, Kropiwnicki E, Wojciechowicz ML, Jagodnik KM, Shu I, Bailey A, Clarke DJB, Jeon M, Evangelista JE, V Kuleshov M, Lachmann A, Parigi AA, Sanchez JM, Jenkins SL, Ma'ayan A. Getting Started with LINCS Datasets and Tools. Curr Protoc. 2022 Jul;2(7):e487. doi: 10.1002/cpz1.487. PMID: 35876555; PMCID: PMC9326873.
- Evangelista JE, Clarke DJB, Xie Z, Lachmann A, Jeon M, Chen K, Jagodnik KM, Jenkins SL, Kuleshov MV, Wojciechowicz ML, Schürer SC, Medvedovic M, Ma'ayan A. SigCom LINCS: data and metadata search engine for a million gene expression signatures. Nucleic Acids Res. 2022 Jul 5;50(W1):W697-W709. doi: 10.1093/nar/gkac328. PMID: 35524556; PMCID: PMC9252724.
- Clarke DJB, Kuleshov MV, Xie Z, Evangelista JE, Meyers MR, Kropiwnicki E, Jenkins SL, Ma'ayan A. Gene and drug landing page aggregator. Bioinform Adv. 2022 Feb 28;2(1):vbac013. doi: 10.1093/bioadv/vbac013. PMID: 35368424; PMCID: PMC8969666.
- Kropiwnicki E, Lachmann A, Clarke DJB, Xie Z, Jagodnik KM, Ma'ayan A. DrugShot: querying biomedical search terms to retrieve prioritized lists of small molecules. BMC Bioinformatics. 2022 Feb 19;23(1):76. doi: 10.1186/s12859-022-04590-5. PMID: 35183110; PMCID: PMC8858480.
- Lachmann A, Xie Z, Ma'ayan A. blitzGSEA: efficient computation of gene set enrichment analysis through gamma distribution approximation. Bioinformatics. 2022 Apr 12;38(8):2356-2357. doi: 10.1093/bioinformatics/btac076. PMID: 35143610; PMCID: PMC9004650.
- Kropiwnicki E, Binder JL, Yang JJ, Holmes J, Lachmann A, Clarke DJB, Sheils T, Kelleher KJ, Metzger VT, Bologa CG, Oprea TI, Ma'ayan A. Getting Started with the IDG KMC Datasets and Tools. Curr Protoc. 2022 Jan;2(1):e355. doi: 10.1002/cpz1.355. PMID: 35085427; PMCID: PMC10789444.
- Piret SE, Attallah AA, Gu X, Guo Y, Gujarati NA, Henein J, Zollman A, Hato T, Ma'ayan A, Revelo MP, Dickman KG, Chen CH, Shun CT, Rosenquist TA, He JC, Mallipattu SK. Loss of proximal tubular transcription factor Krüppel-like factor 15 exacerbates kidney injury through loss of fatty acid oxidation. Kidney Int. 2021 Dec;100(6):1250-1267. doi: 10.1016/j.kint.2021.08.031. Epub 2021 Oct 9. PMID: 34634362; PMCID: PMC8608748.
- Piret SE, Guo Y, Attallah AA, Horne SJ, Zollman A, Owusu D, Henein J, Sidorenko VS, Revelo MP, Hato T, Ma'ayan A, He JC, Mallipattu SK. Krüppel-like factor 6-mediated loss of BCAA catabolism contributes to kidney injury in mice and humans. Proc Natl Acad Sci U S A. 2021 Jun 8;118(23):e2024414118. doi: 10.1073/pnas.2024414118. PMID: 34074766; PMCID: PMC8201852.
- Kuleshov MV, Xie Z, London ABK, Yang J, Evangelista JE, Lachmann A, Shu I, Torre D, Ma'ayan A. KEA3: improved kinase enrichment analysis via data integration. Nucleic Acids Res. 2021 Jul 2;49(W1):W304-W316. doi: 10.1093/nar/gkab359. PMID: 34019655; PMCID: PMC8265130.
- Sevilla A, Papatsenko D, Mazloom AR, Xu H, Vasileva A, Unwin RD, LeRoy G, Chen EY, Garrett-Bakelman FE, Lee DF, Trinite B, Webb RL, Wang Z, Su J, Gingold J, Melnick A, Garcia BA, Whetton AD, MacArthur BD, Ma'ayan A, Lemischka IR. An Esrrb and Nanog Cell Fate Regulatory Module Controlled by Feed Forward Loop Interactions. Front Cell Dev Biol. 2021 Mar 19;9:630067. doi: 10.3389/fcell.2021.630067. PMID: 33816475; PMCID: PMC8017264.
- Kropiwnicki E, Evangelista JE, Stein DJ, Clarke DJB, Lachmann A, Kuleshov MV, Jeon M, Jagodnik KM, Ma'ayan A. Drugmonizome and Drugmonizome-ML: integration and abstraction of small molecule attributes for drug enrichment analysis and machine learning. Database (Oxford). 2021 Mar 31;2021:baab017. doi: 10.1093/database/baab017. PMID: 33787872; PMCID: PMC8011435.
- Xie Z, Bailey A, Kuleshov MV, Clarke DJB, Evangelista JE, Jenkins SL, Lachmann A, Wojciechowicz ML, Kropiwnicki E, Jagodnik KM, Jeon M, Ma'ayan A. Gene Set Knowledge Discovery with Enrichr. Curr Protoc. 2021 Mar;1(3):e90. doi: 10.1002/cpz1.90. PMID: 33780170; PMCID: PMC8152575.
- Clarke DJB, Rebman AW, Bailey A, Wojciechowicz ML, Jenkins SL, Evangelista JE, Danieletto M, Fan J, Eshoo MW, Mosel MR, Robinson W, Ramadoss N, Bobe J, Soloski MJ, Aucott JN, Ma'ayan A. Predicting Lyme Disease From Patients' Peripheral Blood Mononuclear Cells Profiled With RNA-Sequencing. Front Immunol. 2021 Mar 8;12:636289. doi: 10.3389/fimmu.2021.636289. PMID: 33763080; PMCID: PMC7982722.
- Clarke DJB, Jeon M, Stein DJ, Moiseyev N, Kropiwnicki E, Dai C, Xie Z, Wojciechowicz ML, Litz S, Hom J, Evangelista JE, Goldman L, Zhang S, Yoon C, Ahamed T, Bhuiyan S, Cheng M, Karam J, Jagodnik KM, Shu I, Lachmann A, Ayling S, Jenkins SL, Ma'ayan A. Appyters: Turning Jupyter Notebooks into data-driven web apps. Patterns (N Y). 2021 Mar 4;2(3):100213. doi: 10.1016/j.patter.2021.100213. PMID: 33748796; PMCID: PMC7961182.
- Jeon M, Jagodnik KM, Kropiwnicki E, Stein DJ, Ma'ayan A. Prioritizing Pain- Associated Targets with Machine Learning. Biochemistry. 2021 May 11;60(18):1430-1446. doi: 10.1021/acs.biochem.0c00930. Epub 2021 Feb 19. PMID: 33606503.
- Zhang L, Wang Z, Liu R, Li Z, Lin J, Wojciechowicz ML, Huang J, Lee K, Ma'ayan A, He JC. Connectivity Mapping Identifies BI-2536 as a Potential Drug to Treat Diabetic Kidney Disease. Diabetes. 2021 Feb;70(2):589-602. doi: 10.2337/db20-0580. Epub 2020 Oct 16. PMID: 33067313; PMCID: PMC7881868.
- Eberlein J, Davenport B, Nguyen TT, Victorino F, Jhun K, van der Heide V, Kuleshov M, Ma'ayan A, Kedl R, Homann D. Chemokine Signatures of Pathogen- Specific T Cells I: Effector T Cells. J Immunol. 2020 Oct 15;205(8):2169-2187. doi: 10.4049/jimmunol.2000253. Epub 2020 Sep 18. PMID: 32948687; PMCID: PMC7541659.
- Davenport B, Eberlein J, Nguyen TT, Victorino F, van der Heide V, Kuleshov M, Ma'ayan A, Kedl R, Homann D. Chemokine Signatures of Pathogen-Specific T Cells II: Memory T Cells in Acute and Chronic Infection. J Immunol. 2020 Oct 15;205(8):2188-2206. doi: 10.4049/jimmunol.2000254. Epub 2020 Sep 18. PMID: 32948682; PMCID: PMC7541577.
- Kuleshov MV, Stein DJ, Clarke DJB, Kropiwnicki E, Jagodnik KM, Bartal A, Evangelista JE, Hom J, Cheng M, Bailey A, Zhou A, Ferguson LB, Lachmann A, Ma'ayan A. The COVID-19 Drug and Gene Set Library. Patterns (N Y) 2020 Sep 11;1(6):100090. PMID: 32838343
- Wojciechowicz ML, Ma'ayan A. GPR84: an immune response dial? Nature Review Drug Discovery 2020 Jun;19(6):374. PMID: 32494048
- Edwards JJ, Rouillard AD, Fernandez NF, Wang Z, Lachmann A, Shankaran SS, Bisgrove BW, Demarest B, Turan N, Srivastava D, Bernstein D, Deanfield J, Giardini A, Porter G, Kim R, Roberts AE, Newburger JW, Goldmuntz E, Brueckner M, Lifton RP, Seidman CE, Chung WK, Tristani-Firouzi M, Yost HJ, Ma'ayan A, Gelb BD. Systems Analysis Implicates WAVE2 Complex in the Pathogenesis of Developmental Left-Sided Obstructive Heart Defects. JACC Basic to Translational Science 2020 Apr 8;5(4):376-386. PMID: 32368696
- Bartal A, Lachmann A, Clarke DJB, Seiden AH, Jagodnik KM, Ma'ayan A. EnrichrBot: Twitter bot tracking tweets about human genes. Bioinformatics 2020 Jun 1;36(12):3932-3934. PMID: 32277816
- Heitman N, Sennett R, Mok KW, Saxena N, Srivastava D, Martino P, Grisanti L, Wang Z, Ma'ayan A, Rompolas P, Rendl M. Dermal sheath contraction powers stem cell niche relocation during hair cycle regression. Science. 2020 Jan 10;367(6474):161-166. PMID: 31857493
- Duncan A, Heyer MP, Ishikawa M, Caligiuri SPB, Liu XA, Chen Z, Micioni Di Bonaventura MV, Elayouby KS, Ables JL, Howe WM, Bali P, Fillinger C, Williams M, O'Connor RM, Wang Z, Lu Q, Kamenecka TM, Ma'ayan A, O'Neill HC, Ibanez-Tallon I, Geurts AM, Kenny PJ. Habenular TCF7L2 links nicotine addiction to diabetes. Nature 2019 Oct;574(7778):372-377. PMID: 31619789
- Fernandez DM, Rahman AH, Fernandez NF, Chudnovskiy A, Amir ED, Amadori L, Khan NS, Wong CK, Shamailova R, Hill CA, Wang Z, Remark R, Li JR, Pina C, Faries C, Awad AJ, Moss N, Bjorkegren JLM, Kim-Schulze S, Gnjatic S, Ma'ayan A, Mocco J, Faries P, Merad M, Giannarelli C. Single-cell immune landscape of human atherosclerotic plaques. Nature Medicine 2019 Oct;25(10):1576-1588. PMID: 31591603
- Nakahara F, Borger DK, Wei Q, Pinho S, Maryanovich M, Zahalka AH, Suzuki M, Cruz CD, Wang Z, Xu C, Boulais PE, Ma'ayan A, Greally JM, Frenette PS. Engineering a haematopoietic stem cell niche by revitalizing mesenchymal stromal cells. Nature Cell Biology 2019 Apr 15. doi: 10.1038/s41556-019-0308-3. PMID: 30988422
- Wang Z, Lachmann A, Ma'ayan A. Mining data and metadata from the gene expression omnibus. Biophysical Reviews 2019 Feb;11(1):103-110. PMID: 30594974
- Ellis RJ, Wang Z, Genes N, Ma'ayan A. Predicting opioid dependence from electronic health records with machine learning. BioData Mining 2019 Jan 29;12:3. PMID: 30728857
- Mok KW, Saxena N, Heitman N, Grisanti L, Srivastava D, Muraro MJ, Jacob T, Sennett R, Wang Z, Su Y, Yang LM, Ma'ayan A, Ornitz DM, Kasper M, Rendl M. Dermal condensate niche fate specification occurs prior to formation and is placode progenitor dependent. Developmental Cell 2019 Jan 7;48(1):32-48.e5. PMID: 30595537
- Oprea TI, Jan L, Johnson GL, Roth BL, Ma'ayan A, Schürer S, Shoichet BK, Sklar LA, McManus MT. Far away from the lamppost. PLoS Biology 2018 Dec 11;16(12):e3000067. PMID: 30532236
- Torre D, Lachmann A, Ma'ayan A. BioJupies: Automated Generation of Interactive Notebooks for RNA-Seq Data Analysis in the Cloud. Cell Systems 2018 Nov 28;7(5):556-561.e3. PMID: 30447998
- Wang Z, He E, Sani K, Jagodnik KM, Silverstein M, Ma'ayan A. Drug Gene Budger (DGB): An application for ranking drugs to modulate a specific gene based on transcriptomic signatures. Bioinformatics. 2018 Aug 31. doi: 10.1093/bioinformatics/bty763. PMID: 30169739
- Clarke DJB, Kuleshov MV, Schilder BM, Torre D, Duffy ME, Keenan AB, Lachmann A, Feldmann AS, Gundersen GW, Silverstein MC, Wang Z, Ma'ayan A. eXpression2Kinases (X2K) Web: linking expression signatures to upstream cell signaling networks. Nucleic Acids Research 2018 Jul 2;46(W1):W171-W179. PMID: 29800326
- Grimes M, Hall B, Foltz L, Levy T, Rikova K, Gaiser J, Cook W, Smirnova E, Wheeler T, Clark NR, Lachmann A, Zhang B, Hornbeck P, Ma'ayan A, Comb M. Integration of protein phosphorylation, acetylation, and methylation data sets to outline lung cancer signaling networks. Science Signaling 2018 May 22;11(531). pii: eaaq1087. PMID: 29789295
- Lachmann A, Torre D, Keenan AB, Jagodnik KM, Lee HJ, Wang L, Silverstein MC, Ma'ayan A. Massive mining of publicly available RNA-seq data from human and mouse. Nature Communications 2018 Apr 10;9(1):1366. PMID: 29636450
- Torre D, Krawczuk P, Jagodnik KM, Lachmann A, Wang Z, Wang L, Kuleshov MV, Ma'ayan A. Datasets2Tools, repository and search engine for bioinformatics datasets, tools and canned analyses. Scientific Data 2018 Feb 27;5:180023. PMID: 29485625
- Wang Z, Lachmann A, Keenan AB, Ma'ayan A. L1000FWD: fireworks visualization of drug-induced transcriptomic signatures. Bioinformatics. 2018 Jun 15;34(12):2150-2152. PMID: 29420694
- Wang Z, Li L, Glicksberg BS, Israel A, Dudley JT, Ma’ayan A. Predicting age by mining electronic medical records with deep learning characterizes differences between chronological and physiological age. Journal of Biomedical Informatics 2017 Nov 4. pii: S1532-0464(17)30240-X. PMID: 29113935
- Niepel M, Hafner M, Duan Q, Wang Z, Paull EO, Chung M, Lu X, Stuart JM, Golub TR, Subramanian A, Ma'ayan A, Sorger PK. Common and cell-type specific responses to anti-cancer drugs revealed by high throughput transcript profiling. Nature Communications 2017 Oct 30;8(1):1186. PMID: 29084964
- Fernandez NF, Gundersen GW, Rahman A, Grimes ML, Rikova K, Hornbeck P, Ma'ayan A. Clustergrammer, a web-based heatmap visualization and analysis tool for high-dimensional biological data. Scientific Data 2017 Oct 10;4:170151. PMID: 28994825
- Asada N, Kunisaki Y, Pierce H, Wang Z, Fernandez NF, Birbrair A, Ma'ayan A, Frenette PS. Differential cytokine contributions of perivascular haematopoietic stem cell niches. Nature Cell Biology 2017 Mar;19(3):214-223. PMID: 28218906
- Shameer K, Glicksberg BS, Hodos R, Johnson KW, Badgeley MA, Readhead B, Tomlinson MS, O'Connor T, Miotto R, Kidd BA, Chen R, Ma'ayan A, Dudley JT. Systematic analyses of drugs and disease indications in RepurposeDB reveal pharmacological, biological and epidemiological factors influencing drug repositioning. Briefings in Bioinformatics 2017 Feb 15. PMID: 28200013
- Gundersen GW, Jagodnik KM, Woodland H, Fernandez NF, Sani K, Dohlman AB, Ung PM, Monteiro CD, Schlessinger A, Ma'ayan A. GEN3VA: aggregation and analysis of gene expression signatures from related studies. BMC Bioinformatics 2016 Nov 15;17(1):461. PMID: 27846806
- Wang Z, Monteiro CD, Jagodnik KM, Fernandez NF, Gundersen GW, Rouillard AD, Jenkins SL, Feldmann AS, Hu KS, McDermott MG, Duan Q, Clark NR, Jones MR, Kou Y, Goff T, Woodland H, Amaral FM, Szeto GL, Fuchs O, Schüssler-Fiorenza Rose SM, Sharma S, Schwartz U, Bausela XB, Szymkiewicz M, Maroulis V, Salykin A, Barra CM, Kruth CD, Bongio NJ, Mathur V, Todoric RD, Rubin UE, Malatras A, Fulp CT, Galindo JA, Motiejunaite R, Jüschke C, Dishuck PC, Lahl K, Jafari M, Aibar S, Zaravinos A, Steenhuizen LH, Allison LR, Gamallo P, de Andres Segura F, Dae Devlin T, Pérez-García V, Ma'ayan A. Extraction and analysis of signatures from the Gene Expression Omnibus by the crowd. Nature Communications 2016 Sep 26;7:12846. PMID: 27667448
- Wang Z, Clark NR, Ma'ayan A. Drug-induced adverse events prediction with the LINCS L1000 data. Bioinformatics 2016 Aug 1;32(15):2338-45. PMID: 27153606
- Wang Z, Ma'ayan A. An open RNA-Seq data analysis pipeline tutorial with an example of reprocessing data from a recent Zika virus study. F1000Research 2016 Jul 5;5:1574. PMID: 27583132
- Rouillard AD, Gundersen GW, Fernandez NF, Wang Z, Monteiro CD, McDermott MG, Ma'ayan A. The harmonizome: a collection of processed datasets gathered to serve and mine knowledge about genes and proteins. Database (Oxford) 2016 Jul 3;2016. PMID: 27374120
- Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, McDermott MG, Monteiro CD, Gundersen GW, Ma'ayan A. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Research 2016 Jul 8;44(W1):W90-7. PMID: 27141961
- Duan Q, Reid SP, Clark NR, Wang Z, Fernandez NF, Rouillard AD, Readhead B, Tritsch SR, Hodos R, Hafner M, Niepel M, Sorger PK, Dudley JT, Bavari S, Panchal RG, Ma'ayan A. L1000CDS2: LINCS L1000 characteristic direction signatures search engine. NPJ Systems Biology and Applications 2016;2. pii: 16015. PMID: 28413689
- Khan JA, Mendelson A, Kunisaki Y, Birbrair A, Kou Y, Arnal-Estapé A, Pinho S, Ciero P, Nakahara F, Ma'ayan A, Bergman A, Merad M, Frenette PS. Fetal liver hematopoietic stem cell niches associate with portal vessels. Science 2016 Jan 8;351(6269):176-80. PMID: 26634440
- Li L, Cheng WY, Glicksberg BS, Gottesman O, Tamler R, Chen R, Bottinger EP, Dudley JT. Identification of type 2 diabetes subgroups through topological analysis of patient similarity. Science Translational Medicine 2015 Oct 28;7(311):311ra174. PMID: 26511511
- Rouillard AD, Wang Z, Ma'ayan A. Abstraction for data integration: Fusing mammalian molecular, cellular and phenotype big datasets for better knowledge extraction. Computational Biology and Chemistry 2015 Oct;58:104-19. PMID: 26101093
- Gundersen GW, Jones MR, Rouillard AD, Kou Y, Monteiro CD, Feldmann AS, Hu KS, Ma'ayan A. GEO2Enrichr: browser extension and server app to extract gene sets from GEO and analyze them for biological functions. Bioinformatics 2015 Sep 15;31(18):3060-2. PMID: 25971742
- Wang Z, Clark NR, Ma'ayan A. Dynamics of the discovery process of protein-protein interactions from low content studies. BMC Systems Biology 2015 Jun 6;9:26. PMID: 26048415
- Duan Q, Wang Z, Fernandez NF, Rouillard AD, Tan CM, Benes CH, Ma'ayan A. Drug/Cell-line Browser: interactive canvas visualization of cancer drug/cell-line viability assay datasets. Bioinformatics 2014 Nov 15;30(22):3289-90. PMID: 25100688
- Ma'ayan A, Duan Q. A blueprint of cell identity. Nature Biotechnology 2014 Oct;32(10):1007-8. PMID: 25299921
- Ma'ayan A, Rouillard AD, Clark NR, Wang Z, Duan Q, Kou Y. Lean Big Data integration in systems biology and systems pharmacology. Trends in Pharmacological Sciences 2014 Sep;35(9):450-60. PMID: 25109570
- Duan Q, Flynn C, Niepel M, Hafner M, Muhlich JL, Fernandez NF, Rouillard AD, Tan CM, Chen EY, Golub TR, Sorger PK, Subramanian A, Ma'ayan A. LINCS Canvas Browser: interactive web app to query, browse and interrogate LINCS L1000 gene expression signatures. Nucleic Acids Research 2014 Jul;42(Web Server issue):W449-60. PMID: 24906883
